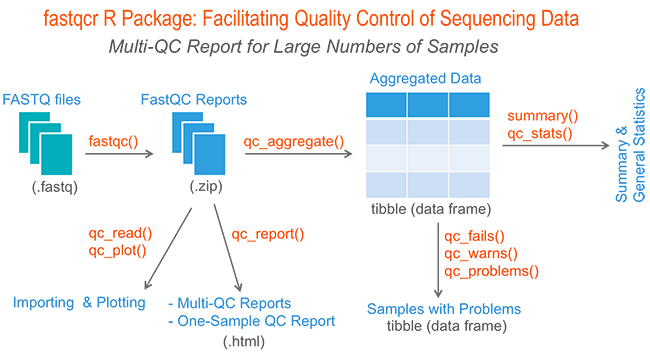
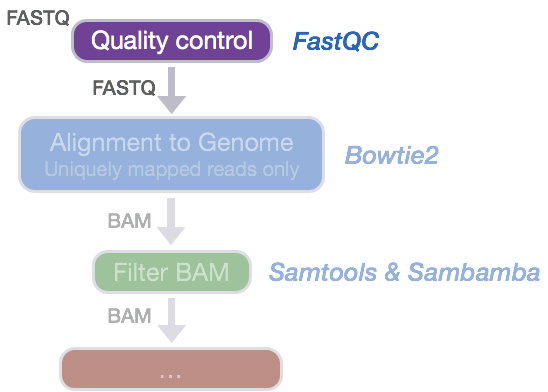
GitHub - natir/fastQt: FastQC port to Qt5: A quality control tool for high throughput sequence data.

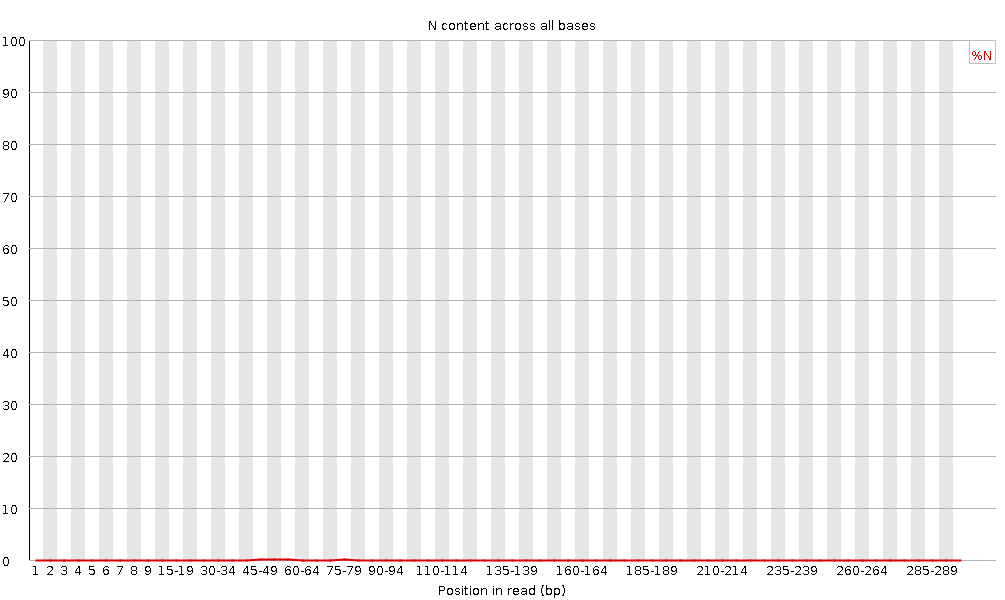
ClinQC quality control report generated by FASTQC. a Per base sequence... | Download Scientific Diagram
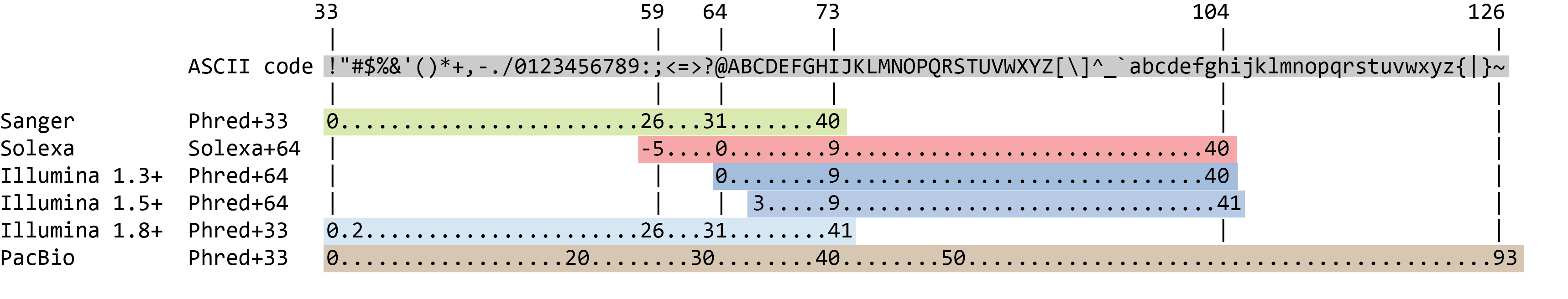
Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data | BibSonomy
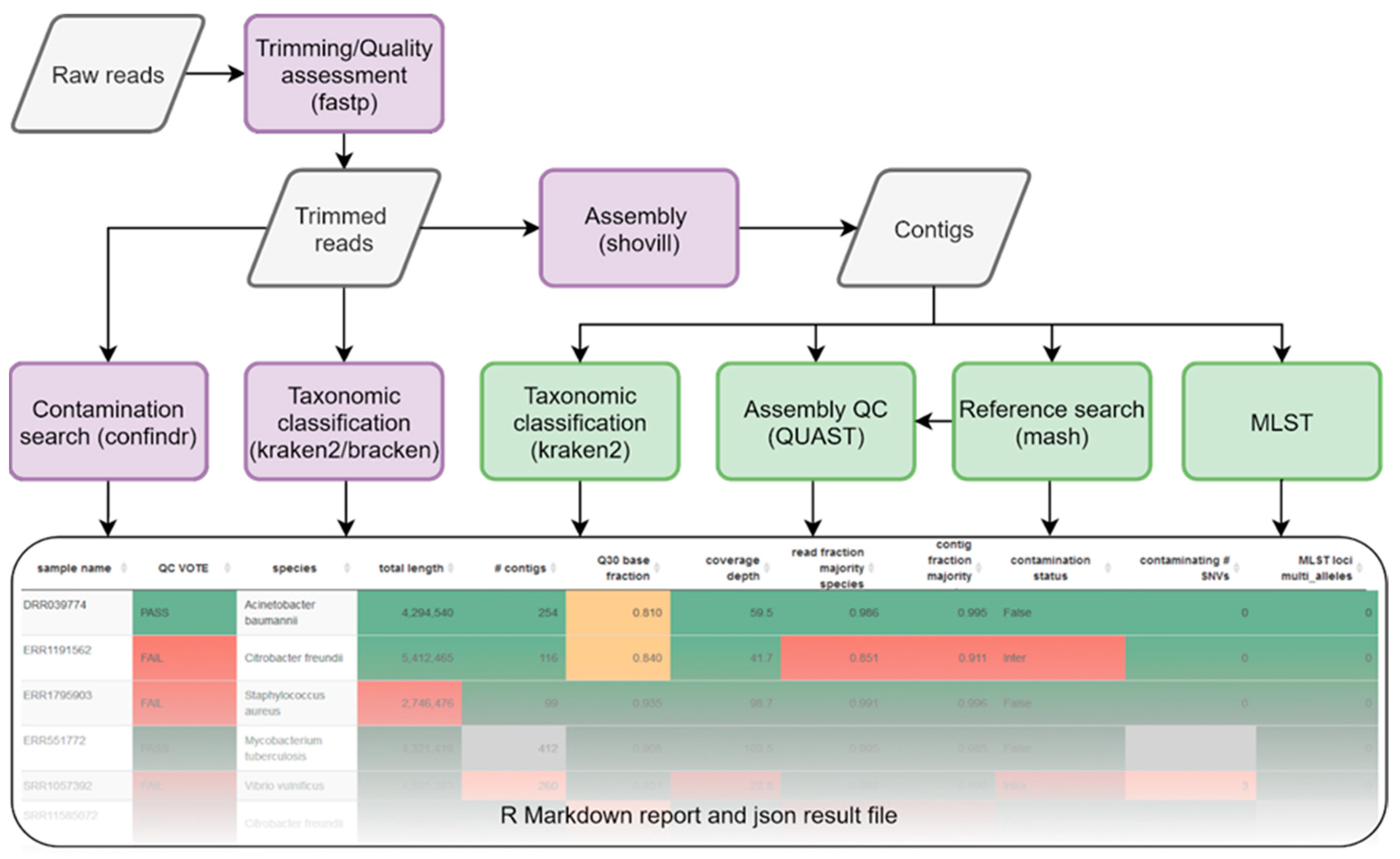
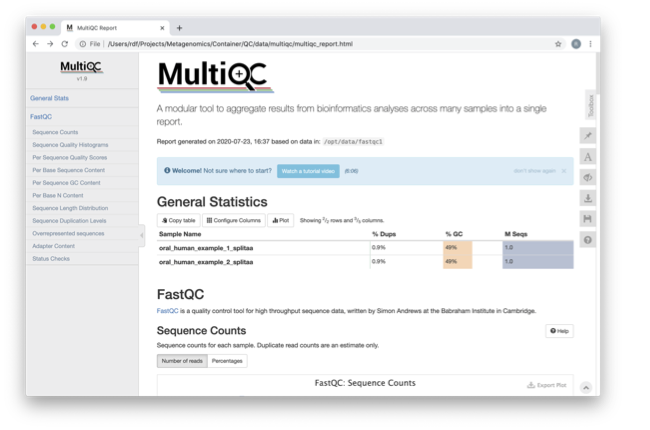
Genes | Free Full-Text | Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with AQUAMIS
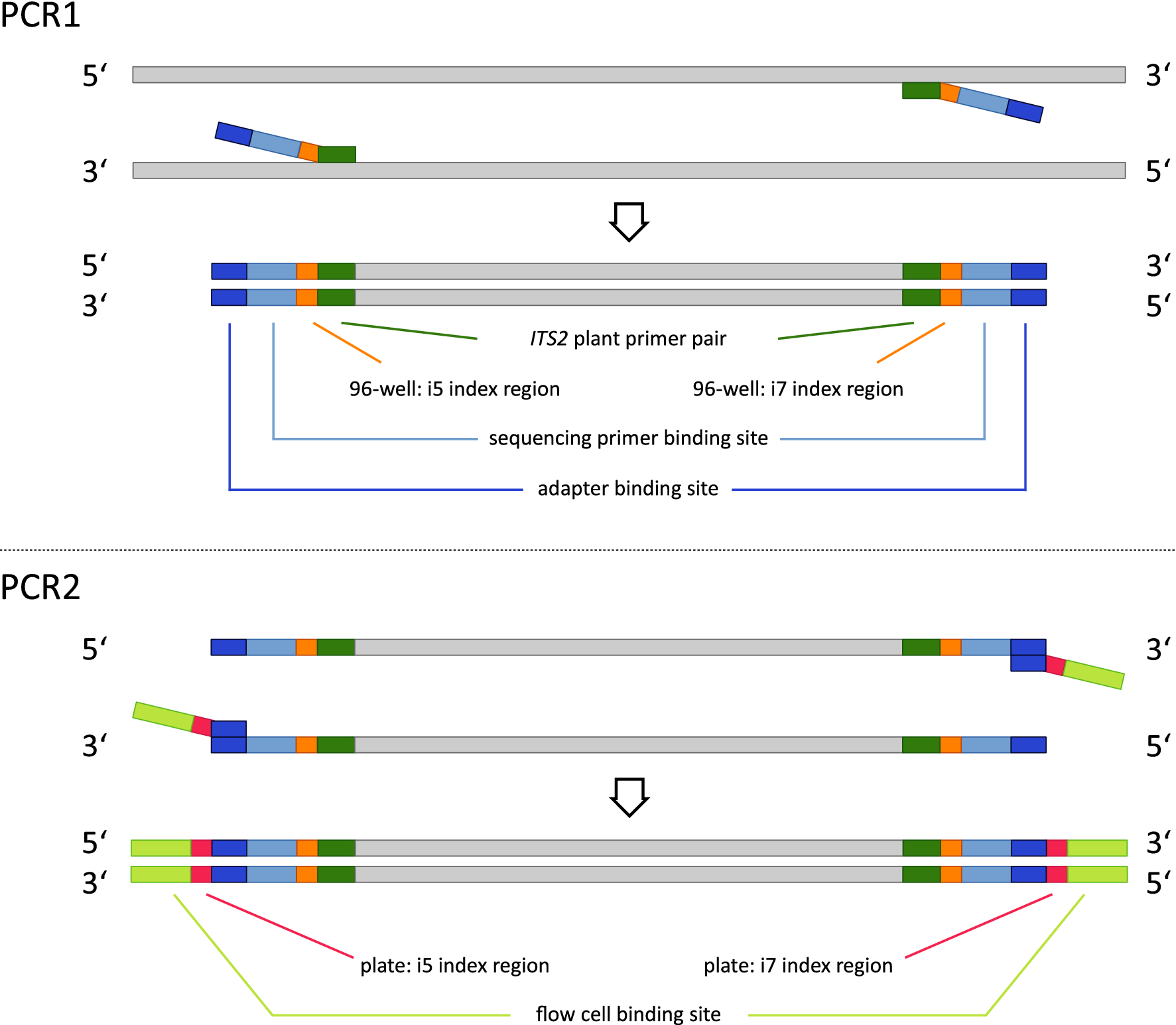
Handling of targeted amplicon sequencing data focusing on index hopping and demultiplexing using a nested metabarcoding approach in ecology | Scientific Reports
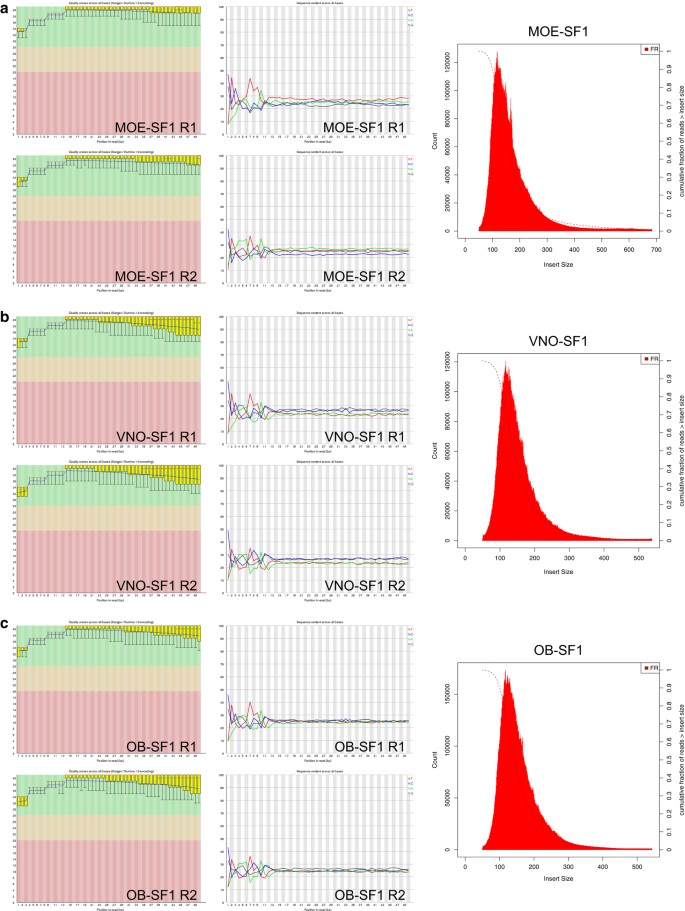
Gene expression profiling of the olfactory tissues of sex-separated and sex-combined female and male mice | Scientific Data

Biomolecules | Free Full-Text | High-Throughput Identification of Adapters in Single-Read Sequencing Data